-Search query
-Search result
Showing all 26 items for (author: chang & js)

EMDB-33393: 
Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs (C1 symmetry)
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33394: 
Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33392: 
Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs at pH ~8.0
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33391: 
Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in GDN detergents at pH ~8.0
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33395: 
Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (GCN conformation)
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33396: 
Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (GCN-TM1i conformation)
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33397: 
Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (FIN conformation)
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33398: 
Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (PLN conformation)
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33399: 
Consensus map of connexin43/Cx43/GJA1 gap junction intercellular channel in LMNG/CHS detergents at pH ~6.9
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS
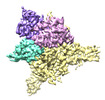
EMDB-27898: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA
Method: single particle / : Johnson ZL, Wasilko DJ, Ammirati M, Chang JS, Han S, Wu H
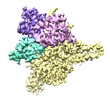
EMDB-27899: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA
Method: single particle / : Wasilko DJ, Johnson ZL, Ammirati M, Chang JS, Han S, Wu H
EMDB-25618: 
SARS-CoV-2 VFLIP spike boung to 2 Ab12 Fab fragments
Method: single particle / : Olmedillas E, Ollmann-Saphire E
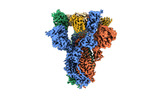
EMDB-25663: 
SARS-CoV-2 S (Spike Glycoprotein) D614G with Three (3) RBDs Up, Bound to Antibody 2-7 scFv, composite map
Method: single particle / : Byrne PO, McLellan JS

EMDB-25689: 
SARS-CoV-2 S (Spike Glycoprotein) D614G with Three (3) RBDs Up, Bound to Antibody 2-7 scFv, global map with poorly-resolved RBDs and scFvs
Method: single particle / : Byrne PO, McLellan JS

EMDB-25690: 
SARS-CoV-2 S (Spike Glycoprotein) D614G with Three (3) RBDs Up, Bound to Antibody 2-7 scFv, local refinement map
Method: single particle / : Byrne PO, McLellan JS

EMDB-25711: 
SARS-CoV-2 S (Spike Glycoprotein) D614G with One(1) RBD Up
Method: single particle / : Byrne PO, McLellan JS

EMDB-31495: 
Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in LMNG/CHS detergents at pH ~8.0
Method: single particle / : Lee HJ, Cha HJ

EMDB-31496: 
Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in nanodiscs with soybean lipids at pH ~8.0
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-31497: 
Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel with two conformationally different hemichannels
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-23521: 
Prefusion RSV F glycoprotein bound by neutralizing site V-directed antibody ADI-14442
Method: single particle / : Gilman MSA, McLellan JS

EMDB-23520: 
Cryo-EM structure of RSV preF bound by Fabs 32.4K and 01.4B
Method: single particle / : Wrapp D, McLellan JS

EMDB-21874: 
Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery
Method: single particle / : Guardia CM, Tan X

EMDB-21876: 
Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery
Method: single particle / : Guardia CM, Tan X

EMDB-21877: 
ATG9A stateA monomer map
Method: single particle / : Guardia CM, Tan X, Lian T

EMDB-21878: 
ATG9A stateB monomer
Method: single particle / : Guardia CM, Tan X, Lian T

EMDB-1869: 
Procapsid of Staphylococcus aureus Pathogenicity Island 1
Method: single particle / : Dearborn AD, Spilman MS, Damle PK, Chang JR, Monroe EB, Saad JS, Christie GE, Dokland T
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
